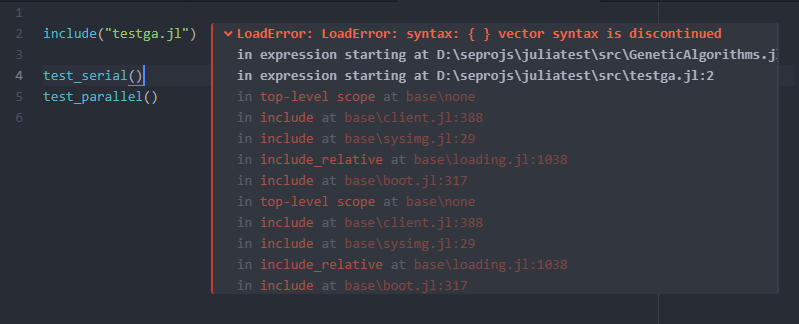
module GeneticAlgorithms
-------
using Base
using Random
export Entity,
GAmodel,
runga,
freeze,
defrost,
generation_num,
population
-------
abstract type Entity end
isless(lhs::Entity, rhs::Entity) = lhs.fitness < rhs.fitness
fitness!(ent::Entity, fitness_score) = ent.fitness = fitness_score
-------
struct EntityData
entity
generation::Int
EntityData(entity, generation::Int) = new(entity, generation)
EntityData(entity, model) = new(entity, model.gen_num)
end
-------
struct GAmodel
initial_pop_size::Int
gen_num::Int
population::Array
pop_data::Array{EntityData}
freezer::Array{EntityData}
rng::AbstractRNG
ga
GAmodel() = new(0, 1, Any[], EntityData[], EntityData[], MersenneTwister(time_ns()), nothing)
end
global _g_model
-------
function freeze(model::GAmodel, entity::EntityData)
push!(model.freezer, entity)
println("Freezing: ", entity)
end
function freeze(model::GAmodel, entity)
entitydata = EntityData(entity, model.gen_num)
freeze(model, entitydata)
end
freeze(entity) = freeze(_g_model, entity)
function defrost(model::GAmodel, generation::Int)
filter(model.freezer) do entitydata
entitydata.generation == generation
end
end
defrost(generation::Int) = defrost(_g_model, generation)
generation_num(model::GAmodel = _g_model) = model.gen_num
population(model::GAmodel = _g_model) = model.population
function runga(mdl::Module; initial_pop_size = 128)
model = GAmodel()
model.ga = mdl
model.initial_pop_size = initial_pop_size
runga(model)
end
function runga(model::GAmodel)
reset_model(model)
create_initial_population(model)
while true
evaluate_population(model)
grouper = @task model.ga.group_entities(model.population)
groupings = Any[]
while !istaskdone(grouper)
group = consume(grouper)
group != nothing && push!(groupings, group)
end
if length(groupings) < 1
break
end
crossover_population(model, groupings)
mutate_population(model)
end
model
end
-------
function reset_model(model::GAmodel)
global _g_model = model
model.gen_num = 1
empty!(model.population)
empty!(model.pop_data)
empty!(model.freezer)
end
function create_initial_population(model::GAmodel)
for i = 1:model.initial_pop_size
entity = model.ga.create_entity(i)
push!(model.population, entity)
push!(model.pop_data, EntityData(entity, model.gen_num))
end
end
function evaluate_population(model::GAmodel)
scores = pmap(model.ga.fitness, model.population)
for i in 1:length(scores)
fitness!(model.population[i], scores[i])
end
sort!(model.population; rev = true)
end
function crossover_population(model::GAmodel, groupings)
old_pop = model.population
model.population = Any[]
sizehint(model.population, length(old_pop))
model.pop_data = EntityData[]
sizehint(model.pop_data, length(old_pop))
model.gen_num += 1
for group in groupings
parents = ( old_pop[i] for i in group )
entity = model.ga.crossover(parents)
push!(model.population, entity)
push!(model.pop_data, EntityData(model.ga.crossover(parents), model.gen_num))
end
end
function mutate_population(model::GAmodel)
for entity in model.population
model.ga.mutate(entity)
end
end
end
这是能够通过的代码(1.0),但是没有测试,需要你自己测试下