我现在就是想跑通作者写得这个代码
(1)(https://github.com/SimonEnsemble/mpn_charges/blob/master/build_graphs/Construct%20graphs%20representing%20the%20MOF.ipynb),花了很长时间,前面都跑通了,现在到最后一块了
@info "making edge list zero-based indexed for Python"
keep_xtal = [true for i = 1:length(xtalnames)]
@showprogress for (i_xtal, xtalname) in enumerate(xtalnames)
crystal = Crystal(xtalname * ".cif", include_zero_charges=true;check_overlap=false)
@assert ! all(crystal.charges.q .== 0)
try
bonds!(crystal, true)
catch e
if isa(e, PyCall.PyError)
remove_bonds!(crystal) # some of the bonds were probz formed
println("Voronoi problems with " * crystal.name)
infer_bonds!(crystal, true) # infer bonds with old distance-based version
else
error("bond! error for " * crystal.name)
end
end
@assert nv(crystal.bonds) == crystal.atoms.n
if ! PorousMaterials.bond_sanity_check(crystal)
keep_xtal[i_xtal] = false
@warn "throwing out " crystal.name
continue
end
###
# node features
# one-hot encoding. each row is an atom.
###
x_ν = zeros(Int, crystal.atoms.n, length(ATOMS))
for (i, atom) in enumerate(crystal.atoms.species)
x_ν[i, ATOM_TO_INT[atom]] = 1
end
@assert sum(x_ν) == crystal.atoms.n
npzwrite(joinpath("graphs1", xtalname * "_node_features.npy"), x_ν)
###
# node labels (the charges)
###
y_ν = deepcopy(crystal.charges.q)
npzwrite(joinpath("graphs1", xtalname * "_node_labels.npy"), y_ν)
@assert length(y_ν) == size(x_ν)[1]
###
# edges
# (a list and their feature = distance btwn atoms)
###
edge_file = open(joinpath("graphs1", xtalname * ".edge_info"), "w")
@printf(edge_file, "src,dst,r\n")
for ed in edges(crystal.bonds)
i = src(ed) # source
j = dst(ed) # destination
r = distance(crystal.atoms, crystal.box, i, j, true)
@printf(edge_file, "%d,%d,%f\n", i - 1, j - 1, r)
end
close(edge_file)
end
writedlm(open("list_of_crystals.txt", "w"), xtalnames[keep_xtal])
Output exceeds the size limit. Open the full output data in a text editor
bond! error for SEHSUU_clean.cif
Stacktrace:
[1] error(s::String)
@ Base .\error.jl:33
[2] macro expansion
@ c:\Users\zzh666\Desktop\任务\任务13-3\111\mpn_charges-master\build_graphs\Construct graphs representing the MOF.ipynb:16 [inlined]
[3] top-level scope
@ C:\Users\zzh666\.julia\packages\ProgressMeter\sN2xr\src\ProgressMeter.jl:938
[4] eval
@ .\boot.jl:360 [inlined]
[5] include_string(mapexpr::typeof(REPL.softscope), mod::Module, code::String, filename::String)
@ Base .\loading.jl:1116
[6] #invokelatest#2
@ .\essentials.jl:708 [inlined]
[7] invokelatest
@ .\essentials.jl:706 [inlined]
[8] (::VSCodeServer.var"#160#161"{VSCodeServer.NotebookRunCellArguments, String})()
@ VSCodeServer c:\Users\zzh666\.vscode\extensions\julialang.language-julia-1.6.24\scripts\packages\VSCodeServer\src\serve_notebook.jl:19
[9] withpath(f::VSCodeServer.var"#160#161"{VSCodeServer.NotebookRunCellArguments, String}, path::String)
@ VSCodeServer c:\Users\zzh666\.vscode\extensions\julialang.language-julia-1.6.24\scripts\packages\VSCodeServer\src\repl.jl:184
[10] notebook_runcell_request(conn::VSCodeServer.JSONRPC.JSONRPCEndpoint{Base.PipeEndpoint, Base.PipeEndpoint}, params::VSCodeServer.NotebookRunCellArguments)
@ VSCodeServer c:\Users\zzh666\.vscode\extensions\julialang.language-julia-1.6.24\scripts\packages\VSCodeServer\src\serve_notebook.jl:13
[11] dispatch_msg(x::VSCodeServer.JSONRPC.JSONRPCEndpoint{Base.PipeEndpoint, Base.PipeEndpoint}, dispatcher::VSCodeServer.JSONRPC.MsgDispatcher, msg::Dict{String, Any})
@ VSCodeServer.JSONRPC c:\Users\zzh666\.vscode\extensions\julialang.language-julia-1.6.24\scripts\packages\JSONRPC\src\typed.jl:67
...
@ Base .\Base.jl:384
[15] exec_options(opts::Base.JLOptions)
@ Base .\client.jl:285
[16] _start()
@ Base .\client.jl:485
(2)
这是bonds自定义函数
function bonds!(crystal::Crystal, apply_pbc::Bool)
if ne(crystal.bonds) > 0
@warn crystal.name * " already has bonds"
end
am = adjacency_matrix(crystal, apply_pbc)
for i = 1:crystal.atoms.n
for j in bonded_atoms(crystal, i, am)
add_edge!(crystal.bonds, i, j)
end
end
end
export bonds!
end
(3)
bonds函数在这个Bonds模块里:(https://github.com/SimonEnsemble/mpn_charges/blob/master/build_graphs/Bonds.jl)
Bonds模块用到scipy包。
这个scipy包我通过一些代码调用的python里的这个scipy包
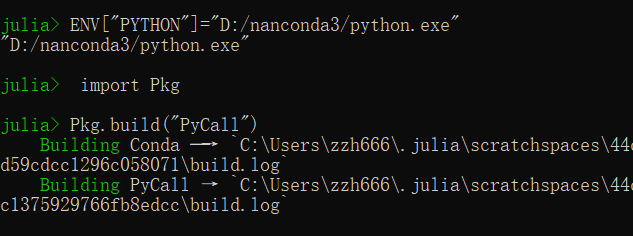
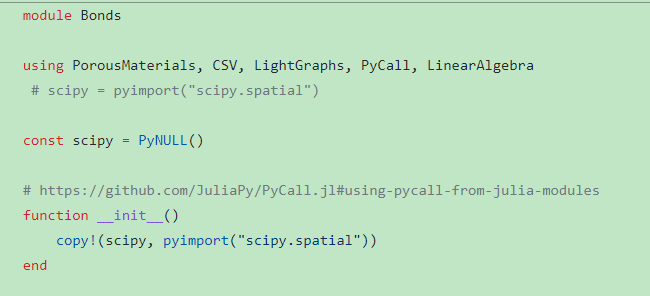
这个错误不知如何解决,不知是不是因为调用scipy的原因,使得Bonds模块不能用,从而bonds函数不能用